|
http://www.abbs.info e-mail:[email protected] ISSN
0582-9879
ACTA BIOCHIMICA et
BIOPHYSICA SINICA 2003, 35(8): 683–688
CN 31-1300/Q |
α-Neurotoxins
of Naja atra and Naja kaouthia Snakes in Different Regions
WEI
Ji-Fu1,2, LÜ
Qiu-Min1, JIN Yang1, Li Dong-Sheng1, XIONG
Yu-Liang1, WANG Wan-Yu1*
( 1Department of
Animal Toxicology, Kunming Institute of Zoology, the Chinese Academy of
Sciences, Kunming 650223, China;2 the Graduate School of the Chinese
Academy of Sciences, Beijing 100039, China )
Abstract Recent studies have shown
that there are geographic variation of α-neurotoxins in Naja kaouthia, but the
cause is not clear yet. In this work, venoms were collected from adult Naja
atra in Zhejiang Province and Naja kaouthia in Yunnan Province, well identified
by morphological characters and cytochrome b gene analysis in summer season to
avoid age and seasonal variation in the venom composition. Then α-neurotoxins
were purified and cloned from these two kinds of snakes. Three α-neurotoxins from Naja kaouthia (Yunnan)
and two from Naja atra (Zhejiang) were identified. Together with previously
reportedα-neurotoxins in Naja kaouthia (Thailand) and Naja atra (Taiwan
Province), it was found that the α-neurotoxins of Naja kaouthia in Yunnan
Province were similar to those of Naja atra in Zhejiang and Taiwan Provinces,
but different from those of Naja kaouthia in Thailand. This result can hardly
be explained by population phylogeny or geographic distance. It might be due to
the different climate, habitat and prey in Thailand in comparison with those in
Yunnan, Zhejiang and Taiwan Provinces.
Key
words Naja kaouthia; Naja
atra; α-neurotoxins; prey
Cobra snakebite is a serious problem in
China. According to Wüster[1], there are two species
of cobras in China, Naja atra and Naja kaouthia. The former is present in most
areas of China including Taiwan Province and the latter is only found in
Southwest China including Yunnan, Sichuan and Guangxi Provinces. Naja kaouthia
is also present in Bihar, Assam, Bengal, Nepal, Indo-China and the Triangle in
Upper Burma.
The major toxic components in cobra
venoms are postsynaptic neurotoxins orα-neurotoxins which block the nerve
transmission by binding specifically to the nicotinic acetylcholine receptor, leading
to flaccid paralysis and even death by respiratory failure. Based on the amino
acid sequences, α-neurotoxins can be divided into two major groups, the long
(65-72 residues with 5 disulfide
bridges) and short neurotoxins (60-62
residues with 4 disulfide bridges). In spite of the diversity in their primary
structure, these two kind of α-neurotoxins share a common three-loop
structure[2,3]. Phylogenetic analysis showed that they were probably derived
from a common ancestor[4]. Cobrotoxin is the main α-neurotoxin found in Taiwan
Naja atra venom. It contains 62 amino acid residues in a single polypeptide
chain[5]. In 1997, another short neurotoxin called cobrotoxin b was purified
from the same source[6]. However, the major α-neurotoxin in Naja kaouthia (Thailand)
venom is a long neurotoxin, α-cobratoxin, the minor α-neurotoxin is different
from cobrotoxin in one residue[7]. Recently, we have purified and sequenced
three short neurotoxins from the venom of Yunnan Naja kaouthia, one being a
novel short neurotoxin, and the other two the same as cobrotoxin and
cobrotoxin-b[8,9]. So, it seemed that there existed geographic variation of
α-neurotoxins in Naja kaouthia. However, the geographic variation of
α-neurotoxins in Naja atra was not reported till now.
The systematics of Asian Naja is in a
confusion. Many problems are due to the fact that these snakes are often
extremely variable even within populations, especially in their coloration and
pattern. This variation has often made the identification extremely difficult,
as snakes in one population may look more different from one another than from
those of another population thousands of miles away[1,10].
To avoid species misidentification, we
collected Zhejiang Naja atra and Yunnan Naja kaouthia, well identified by
morphological characters and partial cytochrome b gene analysis. Venoms were
collected from adult snakes of these two species of cobra in summer season to
avoid age and seasonal variation in the venom composition. Then α-neurotoxins
were purified from these two venoms. On the other hand, α-neurotoxins were also
cloned from the same snake. The similarity and variation of α-neurotoxins
between these venoms were discussed.
1 Materials
and Methods
1.1 Materials
Yunnan Naja kaouthia venom was collected
from Wenshan County, South Yunnan Province, China. Zhejiang Naja atra venom was
collected from Jinghua City, Zhejiang Province, China. SP-Sephadex C-25,
Superdex-75, endoproteinase Glu-C and low-molecular-weight markers were from
Pharmacia Fine Chemical, Uppsala, Sweden. Rats (180-200 g) were from Animal Center, Kunming
Institute of Zoology, CAS, China. Other chemicals and reagents were of
analytical grade.
1.2 Isolation of α-neurotoxins
α-neurotoxins from Zhejiang Naja atra and
Yunnan Naja kaouthia venoms were isolated according to the procedure of Lu et
al.[8]. The lyophilized venom (5 g) was dissolved in 20 mL 0.05 mol/L ammonium
acetate buffer (pH 5.8) and applied to a SP-Sephadex C-25 column (5 cm×60 cm) pre-equilibrated with the same
buffer. The adsorbed proteins were eluted with a linear gradient of 0-1 mol/L NaCl. Separation on Superdex-75
column (1.6 cm×40 cm) was performed with the
same buffer containing 0.15 mol/L NaCl. The sample was then loaded on a
Resource S ion-exchange column (Pharmacia Fine Chemical, Uppsala, Sweden).
Finally, the toxins were purified on HPLC Nava-Pak C18 column (Dalian Elite
Scientific Instruments Co., Ltd., Dalian, China)
1.3 Mass spectrometry
The mass spectra of the derivatives of
isolated components above, their derivatives were recorded on a Bruker reflex
III (Bruker) spectrometer, using α-cyano-4-hydroxycinnamic acid and
2,5-dihydroxy-benzoic acid as matrices.
1.4 Protein sequence analysis
Amino acid sequencing was carried out
with an Applied Biosystem 476A protein sequencer. The reduced and
S-carboxymethylated (RCM-) protein was subjected to automated Edman degradation
to determine the N-terminal sequence. The RCM-protein was hydrolyzed with Glu-C
protease. The hydrolysates were separated by HPLC on a Nava-Pak C18 column (3.9
mm×30 mm). The amino acid
sequence of each peptide fraction was determined.
1.5 Assay of neurotoxicity
Neurotoxicity was assayed according to
the method of Cai et al.[11] Briefly, rats of either sex, weighing between 180-200 g were used. The diaphragm and nervus
phrenicus preparation was quickly excised. The preparation was mounted in a 50
mL organ bath with the diaphragm and nervus phrenicus connected to two separate
electrodes. After the preparation was equilibrated for 60 min in aerated (95%
O2 and 5% CO2) Krebs’ solution, components of different
concentrations were added to the bath to inhibit contractions induced by
electric stimulation.
1.6 Molecular cloning of short neurotoxins
Isolation of mRNA and reverse
transcription were conducted using PolyATract system 1000-kit and Reverse
transcription system kit (Promega Biotech) respectively, according to the
manufacturer’s protocols. Two oligonucleotide primers, designed according to
the signal peptide and 3'-noncoding regions of cobrotoxin gene with the forward
sequence, 5'-ATGAAAACTCTGCTGACCTTGGTG-3' and the reverse one,
5'-GGATGGTCCTTGATGGATGAGAG-C-3', were synthesized[12].
PCR was carried out in 100 μL reaction buffer using total RT-PCR
products as templates. The amplification was processed on a thermocycler 94 ℃/55 ℃/72
℃ 1 min each. The recovered PCR
products were cloned into pMD18-T vector (TaKaRa, Dalian, China) according to
the TA-cloning procedures, and then transformed into E. coli strain JM
109.
The white transformants were screened by
PCR using the above primers, and the positive clones were sequenced in TaKaRa
Biotechnology Co. Ltd. (Dalian, China).
1.7 Amplification and sequencing of partial sequence of cytochrome b
DNA extractions were performed by first
digesting snake liver tissues for overnight at 55 ℃ in 2 mL lyses buffer (100 mmol/L
Tris-HCl, pH 8.0; 50 mmol/L EDTA; 10 mmol/L NaCl; 0.5% SDS) containing
proteinase K at a concentration of 0.06 g/L. Further digesting was carried out
in the same lyses buffer for approximately 3 h at 65 ℃ with constant motion, followed by two
times of extraction in chloroform. Then DNA was precipitated with ethanol and
washed with 80% ethanol. The precipitated DNA was dissolved with TE buffer and
diluted to an appropriate concentration (100-400
mg/ L) prior to PCR. A 767 bp fragment of the cytochrome b gene was amplified
by PCR reaction according to Slowinski et al.[13].
2 Results
2.1 Purification of α-neurotoxins
As shown in Fig.1(A), the Yunnan Naja
kaouthia venom was separated into several protein peaks in SP-Sephadex C-25
chromatography. The peak VI, peak VII and peak VIII were with neurotoxin
activity. Three neurotoxins named as NT-I, NT-II, NT-III were purified from
these peaks according to Lu et al.[8]. The Zhejiang Naja atra venom was
separated with the same procedure, and the peak 6 and 8 were with neurotoxin
activity as shown in Fig.1(B).
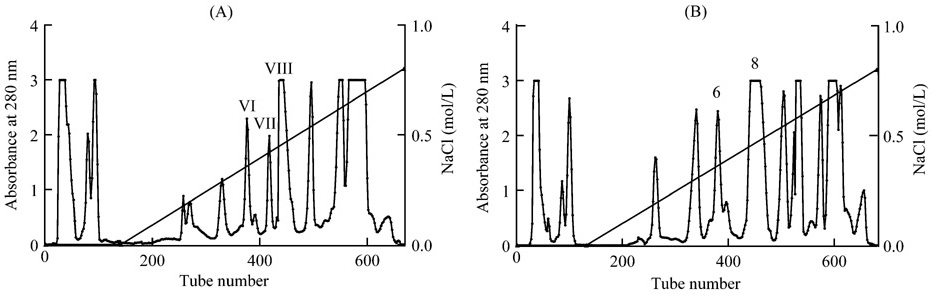
Fig.1 Separation of Naja kaouthia and Naja atra venom on SP-Sephadex C-25 column
(A) Separation of Yunnan Naja kaouthia venom. Peaks VI, VII and VIII contained neurotoxin activity. (B) Separation of Zhejiang Naja atra venom. Peaks 6 and 8 contained neurotoxin activity.
The peak 6 of Zhejiang Naja atra was
further loaded on a Superdex-75 column (1.6 cm×40
cm), and peak 6-3 with neurotoxicity was pooled (Fig.2).
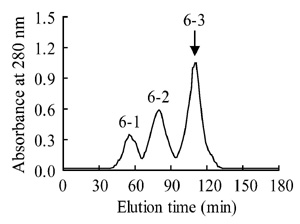
Fig.2 Further separation of the peak 6 in Fig.1(B) on a Superdex-75 column (1.6 cm×40 cm)
The peak containing neurotoxin activity was indicated by arrow.
The peak 8 and the peak 6-3 were loaded
on FPLC Resource S column respectively (Fig.3). The two neurotoxins were
further purified by HPLC Nava-Pak C18 column (Fig.4) and named as NT-I' and
NT-III'. Their molecular weight were 6952.84 and 6829.07 respectively by
matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS.
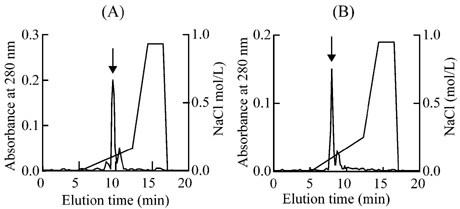
Fig.3Purification of the
neurotoxins of Zhejiang Naja atra on FPLC Resource S column
(A) Separation of NT-I'. (B) Separation
of NT-III′. The peaks containing
neurotoxin activity were indicated by arrows.
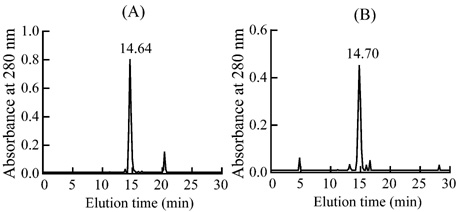
Fig.4 Purification of the NT-I′ and NT-III′ of Zhejiang Naja atra on HPLC Nava-Pak C18 column
(A) Separation of NT-I′. (B) Separation of NT-III′. Retention times of the neurotoxins were marked on the top of the peaks.
2.2 Protein sequence analysis
The two neurotoxins NT-I' and NT-III'
from Zhejiang Naja atra were subjected to amino acid sequencer to determine the
N-terminal sequence. The N-terminal sequences up to 40 amino acids were
determined. Furthermore, the two neurotoxins were subjected to reduction and
S-carboxymethylation. The RCM-neurotoxins were then digested with Glu-C
protease. The hydrolysates of NT-I' and NT-III' were separated into four and
three peptide fractions on Nava-Pak C18 column, respectively. The sequences of
hydrolysates of NT-I' were: INCCTTDRCNN, RGCGCPSVKNGIE, TNCYKKRWRDHRGYRTE and
LECHNQQSSQTPTTTGCSGGE. The sequences of hydrolysates of NT-III' were:
TNCYKKWWSDHRG-TIIE, LECHNQQSSQTPTTKTCSGE and RGCGCPK-VKPGVNLNCCTTDRCNN.
2.3 cDNA cloning of short neurotoxins of Yunnan Naja kaouthia and Zhejiang Naja atra
PCR amplification using venom cDNA
mixtures as templates with the designed primers resulted in a PCR fragment
estimated to be about 300 bp (data not shown). The PCR fragments were then
subcloned by TA-cloning kit. More than 30 clones were selected for sequencing.
The cDNAs of NT-I, NT-II and NT-III from Naja kaouthia and NT-I', NT-III' from
Naja atra were identified, and it was found that the cDNA sequences of NT-I and
NT-III were the same as NT-I' and NT-III' respectively. Combined with the
sequences of hydrolysates of NT-I' and NT-III', the whole amino acid sequences
of NT-I' and NT-III' were determined (Fig.5). Sequence analysis also showed
that the nucleosides and amino acid sequences of NT-I, NT-I' and NT-III,
NT-III' were the same as those of cobrotoxin and cobrotoxin-b, respectively.

Fig.5 Complete cDNA sequences and deduced amino acid sequences of NT-I(NT-I′), NT-II, NT-III(NT-III′)
The nucleotide residues of coding region were numbered in the 5′ to 3′ direction. Beneath the nucleotide sequence is the deduced amino acid sequence (in bold).
2.4 Analysis of partial sequence of cytochrome b
A 767 bp fragment of the cytochrome b
gene was amplified by PCR reaction and sequenced (data not shown). According to
Slowinski et al.[13], Naja atra and Naja kaouthia had the closest evolutional
relationship compared with other cobras. The partial sequence of cytochrome b
of Thailand Naja kaouthia had 93% similarity with that of Taiwan Naja atra.
However, the similarity of partial sequence of cytochrome b between Thailand
and Yunnan Naja kaouthia, Zhejiang and Taiwan Naja atra, were 97% and 99%,
respectively. It indicated there was no possibility of species misidentify.
3 Discussion
Variation in snake venom composition is a
ubiquitous phenomenon at all taxonomic levels. Many factors including
phylogeny, geographic origin, season, age and prey preference may influence
venom composition. Recently, Mukherjee et al.[14] found that there existed high
variations in two closely related snakes, Naja kaouthia and Naja naja collected
in the same place and in the venom composition of Naja naja from three
neighboring districts of India excluding age, sex and seasonal
variation[14,15]. In this work, we first collected the male snakes with almost
the same size. Morphological characters and cytochrome b gene analysis of
Zhejiang Naja atra and Yunnan Naja kaouthia were undertaken to avoid species
misidentification. Snake venoms were collected in summer to eliminate the
influence of season. Results showed that the two venoms from Yunnan Naja
kaouthia and Zhejiang Naja atra differed in chromatographic elution profile
through the identical SP-Sephadex C-25 column. Moreover, NT-II in the former
venom was not found in the latter. Both venoms contained two short neurotoxins
which are identical to cobrotoxin and cobrotoxin-b respectively. In both venoms,
no components with molecular weight similar to that of α-cobratoxin were
detected in the peaks with neurotoxicity by mass spectrometry (data not shown).
However, Yunnan Naja kaouthia venom was distinct from that of Thailand.
α-cobratoxin, the main α-neurotoxin of Thailand Naja kaouthia, was not
found in Yunnan Naja kaouthia. It seemed that intraspecies variation of
α-neurotoxins in Naja kaouthia venom was larger than interspecies variation
between Naja atra and Naja kaouthia. It can hardly be explained by population
phylogeny or geographic distance (Yunnan is nearer to Thailand than to Zhejing
or Taiwan).
In recent years, intraspecies variation
of certain components in snake venoms has received considerable attention[16-18]. On the other hand, there are also
reports that different snake species venoms contained same or similar
components. Tsai et al.[17] found two PLA2s designated as CRV-R6a and CRV-R6b,
cloned from the venom of Calloselasma rhodostoma, had a structure identical to
that of TmPL-III, a PLA2 from Trimeresurus mucro-squamatus, although
Trimeresurus mucrosquamatus and Calloselasma rhodostoma were only loosely
related in evolution. The Lys-49 PLA2 from Trimeresurus mucrosquamatus venoms
was also identical to that from Deinagkistrodon acutus[19]. Cytotoxin 3,
cytotoxin 10 and CX1-NAJSP of Naja atra[20], Thailand Naja kaouthia[21] and
Naja sputatrix[22] had the same sequence. So the variation of purified
components in the same snake venom or similarity of purified components in
different snake venoms is not an unusual phenomenon.
Daltry et al.[23] demonstrated a
significant relationship between geographical variation and venom composition,
and hypothesized that geographical variation in venom composition may reflect
natural selection for higher efficiency in killing and /or digesting different
preys in different regions. The following evidences implied that the
composition of snake venom was strongly influenced by environmental factors
including habitat, climate and preys[16,24]. The habitat, climate of Wenshan
(South Yunnan), Jinhua (South Zhejiang) and Taiwan are similar in summer, but
different from those in Thailand.
Cobras were reported to have a broad diet
spectrum including small mammals, frogs, birds and lizards. So, its diet
analysis in different regions is laborious, expensive and hazardous, and it’s
very hard to get the true all-round diet. Small mammals especially Mus and
Rattus genus are the main prey of Naja atra and Naja kaouthia[25]. So statistic
analysis of Mus and Rattus genus suitable for cobras’ feeding in different
regions could reflect their rough diets composing. We found that four Mus and
Rattus genus in Zhejiang and Yunnan, five in Taiwan, and eleven in Thailand
were the possible prey of cobras[26] , which showed that Thailand was greatly
different from Zhejiang, Yunnan and Taiwan Provinces in diet of cobras.
Long neurotoxins and short neurotoxins
might coexist in the common ancestor of Naja atra and Naja kaouthia since there
existed both long neurotoxins and short neurotoxins in modern Naja kaouthia and
other cobra venoms[4,7], but then evolved independently since there was no
intercourse between their genes. Even in the same species, if two populations
in the different area with different habitats have been separated from each
other for long time in the geological history, namely there was a vicariance
between them, it was certain that the two different populations would vary from
each other. Due to the isolation of Trans-Himalayas Mountains and four big
rivers (Irrowardi River, Saween River, Meigong River and Jinsha River), Naja
kaouthia snakes can be divided to two populations. One is distributed in Bengal
and Northern Berma, and the other in Southern Yunnan, Southwestern Sichuan and
Southeastern Guangxi in China. These two populations had different habitat,
climate and prey, so that it was very possible that they varied from each
other, which could explain the variation of α-neurotoxins in Thailand and
Yunnan Naja kaouthia venom. Though there was also a geographical obstacle
(Taiwan Strait) between Zhejiang and Taiwan Naja atra snakes, they shared
similar habitat, climate and prey, so they contained same α-neurotoxins. This
hypothesis could further explain the similarity of α-neurotoxins between Yunnan
Naja kaouthia and Zhejiang Naja atra. Such cobras populations as those of South
Yunnan and Zhejiang that shared similar habitat, climate and prey might contain
similar α-neurotoxins after the divergence of Naja kaouthia and Naja atra from
the common ancestor.
References
1 Wüster W. Taxonomic changes and toxicology:
Systematic revisions of the Asiatic cobras (Naja naja species complex).
Toxicon, 1996, 34: 399-406
2 Yang CC. Structure and
function of cobra neurotoxin. Adv Exp Med Biol, 1996, 391: 85-96
3 Tsetlin V. Snake venom
alpha-neurotoxins and other ‘three-finger’ proteins. Eur J Biochem, 1999, 264:
281-286
4 Chang L, Lin S, Wang J,
Hu WP, Wu B, Huang H. Structure-function studies on Taiwan cobra long
neurotoxin homolog. Biochim Biophys Acta, 2000, 1480: 293-301
5 Yang CC. Cobrotoxin:
Structure and function. J Nat Toxins, 1999, 8: 221-332
6 Chang LS, Chou YC, Lin
SR, Wu BN, Lin J, Hong E, Sun YJ et al. A novel neurotoxin, cobrotoxin b, from
Naja naja atra (Taiwan cobra) venom: Purification, characterization, and gene
organization. J Biochem (Tokyo), 1997, 122: 1252-1259
7 Chiou SH, Lin WW, Chang
WP. Sequence characterization of venom toxins from Thailand cobra. Int J Pept
Protein Res, 1989, 34: 148-152
8 Lu QM, Meng QX, Li DS, Zhu
SW, Jia YH, Xiong YL, Wang WY. Comparative study of three short-chain
neurotoxins from the venom of Naja kaouthia (Yunnan, China). J Nat Toxins,
2002, 11: 221-229
9 Meng QX, Wang WY, Lu
QM, Jin Y, Wei JF, Zhu SW, Xiong YL. A novel short neurotoxin, cobrotoxin c,
from monocellate cobra (Naja kaouthia) venom: Isolation and purification,
primary and secondary structure determination, and tertiary structure modeling.
Comp Biochem Physiol C Toxicol Pharmacol, 2002, 132: 113-121
10 Wüster W, Golay P, Warrell DA. Synopsis of
recent developments in venomous snake systematics. Toxicon, 1997, 35: 319-340
11Cai
JX, Sun X, Yang CJ, Chen XL, Li CD, Zhao QK, Tian YF et al. Column
chromatography fractionation of ophiophagus hannah venom and study of
neurotoxic peaks. Zoological Reasearch, 1980, 1: 319-325
12 Chang LS, Lin J, Chou YC,
Hong E. Genomic structures of cardiotoxin 4 and cobrotoxin from Naja naja atra
(Taiwan cobra). Biochem Biophys Res Commun, 1997, 239: 756-762
13 Slowinski JB, Keogh JS.
Phylogenetic relationships of elapid snakes based on cytochrome b mtDNA
sequences. Mol Phylogenet Evol, 2000, 15: 157-164
14 Mukherjee AK, Maity CR.
Biochemical composition, lethality and pathophysiology of venom from two cobras——Naja naja and N. kaouthia. Comp Biochem
Physiol B Biochem Mol Biol, 2002, 131: 125-132
15 Mukherjee AK, Maity CR. The
composition of Naja naja venom samples from three districts of West Bengal,
India. Comp Biochem Physiol A Mol Integr Physiol, 1998, 119: 621-627
16 Chijiwa T, Deshimaru M,
Nobuhisa I, Nakai M, Ogawa T, Oda N, Nakashima K et al. Regional evolution of venom-gland phospholipase A2
isoenzymes of Trimeresurus flavoviridis snakes in the southwestern islands of
Japan. Biochem J, 2000, 347: 491-499
17 Tsai IH, Chen YH, Wang YM,
Liau MY, Lu PJ. Differential expression and geographic variation of the venom
phospholipases A2 of Calloselasma rhodostoma and Trimeresurus mucrosquamatus.
Arch Biochem Biophys, 2001, 387: 257-264
18 Wei JF, Wei Q, Lu QM, Tai H, Jin
Y, Wang WY, Xiong YL. Purification, characterization and biological activity of
an L-amino acid oxidase from Trimeresurus mucrosquamatus venom. Acta Biochim
Biophys Sin, 2003, 35: 219-224
19 Wang YM, Wang JH, Pan FM,
Tsai IH. LYS-49 phospholipase A2 homologs from venoms of Deinagkistrodon acutus
and Trimeresurus mucrosquamatus have identical protein sequence. Toxicon, 1996,
34: 485-489
20 Chang LS, Huang HB, Lin SR.
The multiplicity of cardiotoxins from Naja naja atra (Taiwan cobra) venom.
Toxicon, 2000, 38: 1065-1076
21 Joubert FJ, Taljaard N. The
complete primary structures of three cytotoxins (CM-6, CM-7 and CM-7A) from
Naja naja kaouthia (Siamese cobra) snake venom. Toxicon, 1980, 18: 455-467
22 Jeyaseelan K, Armugam A,
Lachumanan R, Tan CH, Tan NH. Six isoforms of cardiotoxin in Malayan spitting
cobra (Naja naja sputatrix) venom: Cloning and characterization of cDNAs.
Biochim Biophys Acta, 1998, 1380: 209-222
23 Daltry JC, Wuster W, Thorpe
RS. Diet and snake venom evolution. Nature, 1996, 379: 537-540
24 Jorge da Silva N Jr, Aird SD.
Prey specificity, comparative lethality and compositional differences of coral
snake venoms. Comp Biochem Physiol C Toxicol Pharmacol, 2001, 128: 425-456
25 Zhao EM. Amphibia and
Reptilia. In: Wang S ed. China Red Data Book of Endangered Animals, Beijing:
Science Press, 1998, 274-279
26 Corbet GB, Hill JE ed. The Mammals
of the Indomalayan Region: A Systematic Review, New York: Oxford University
Press, 1992, 320-413
_______________________________________
Received:
March 31, 2003 Accepted:
May 18, 2003
This
work was supported by a grant from the National Natural Science Foundation of
China (No. 39670165)
*Corresponding
author: Tel, 86-871-5192476; Fax, 86-871-5191823; e-mail, [email protected]